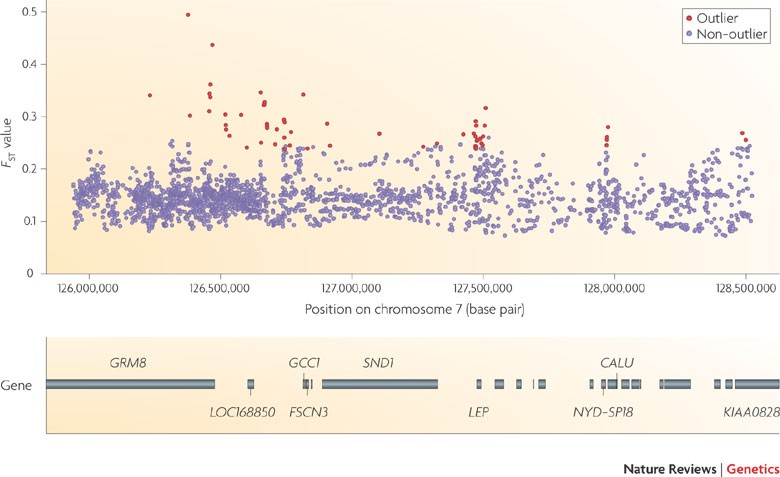
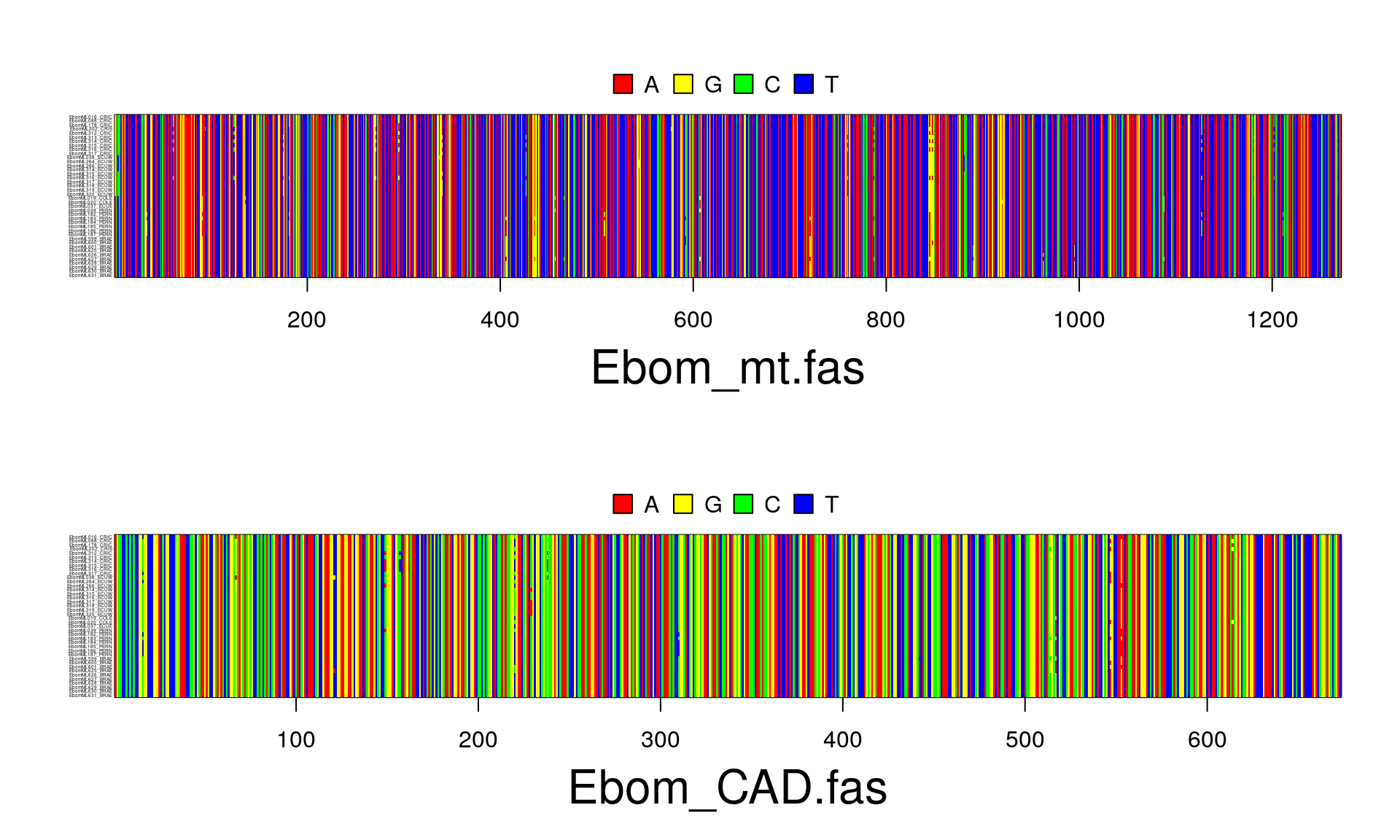
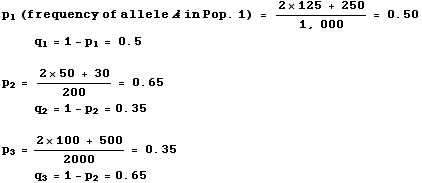
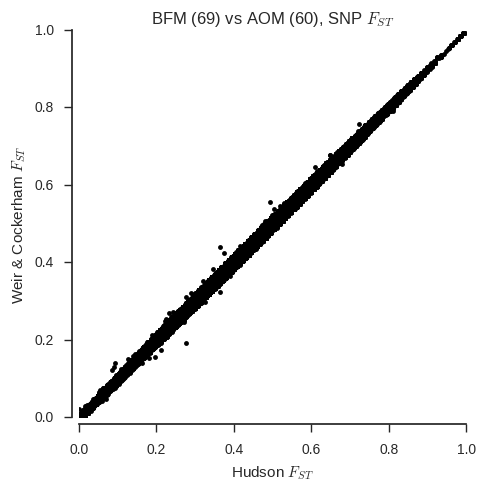
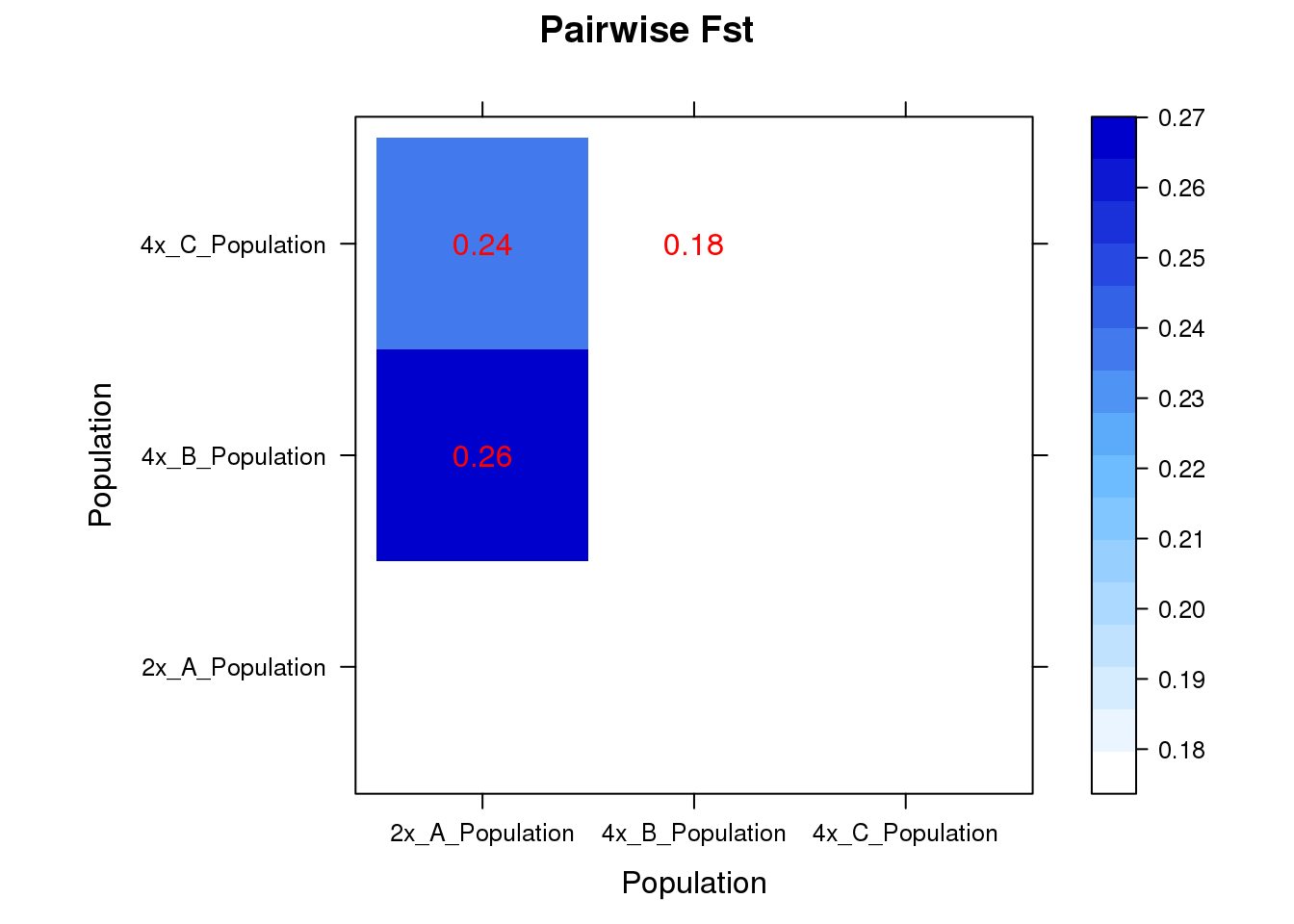
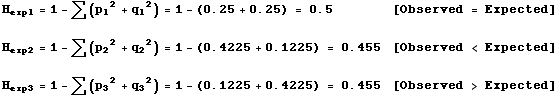SambaR: an R package for fast, easy and reproducible population-genetic analyses of biallelic SNP datasets | bioRxiv
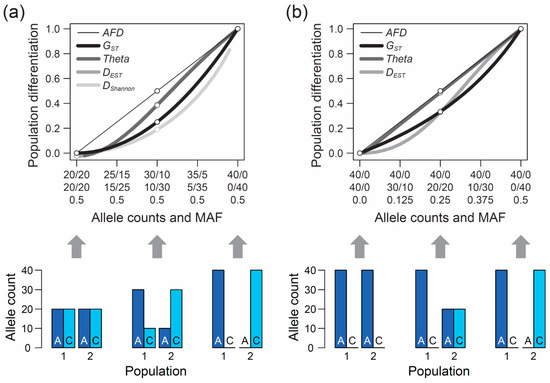
Genes | Free Full-Text | Allele Frequency Difference AFD–An Intuitive Alternative to FST for Quantifying Genetic Population Differentiation

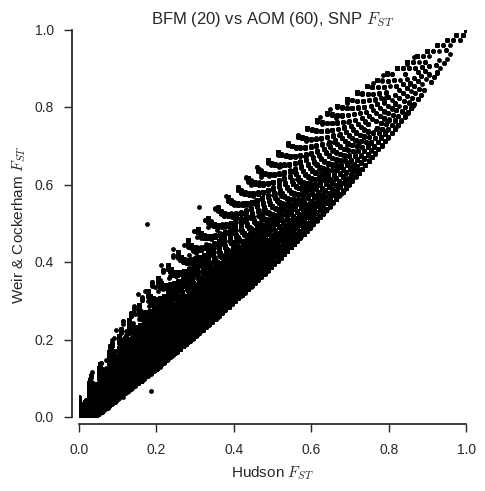
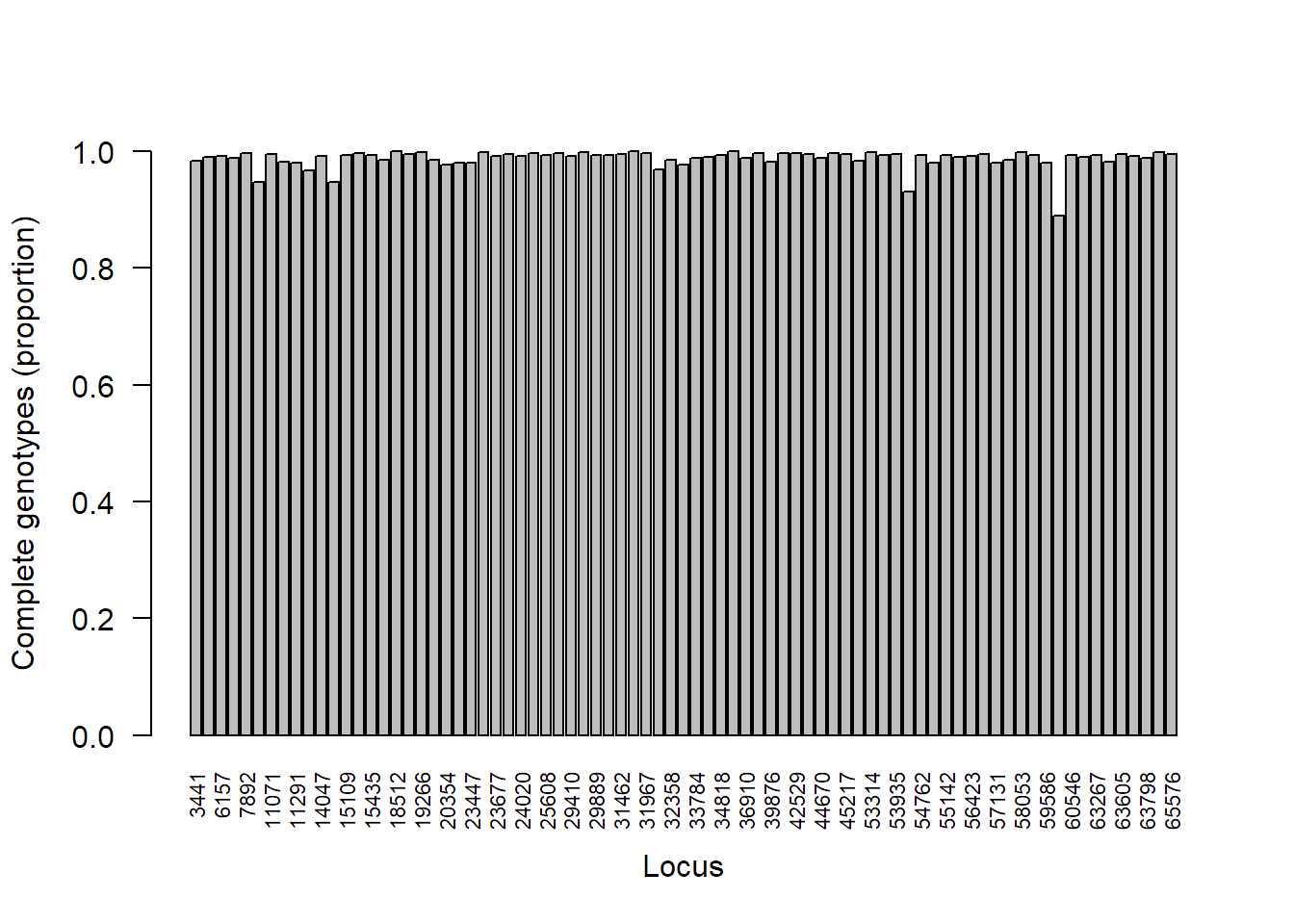
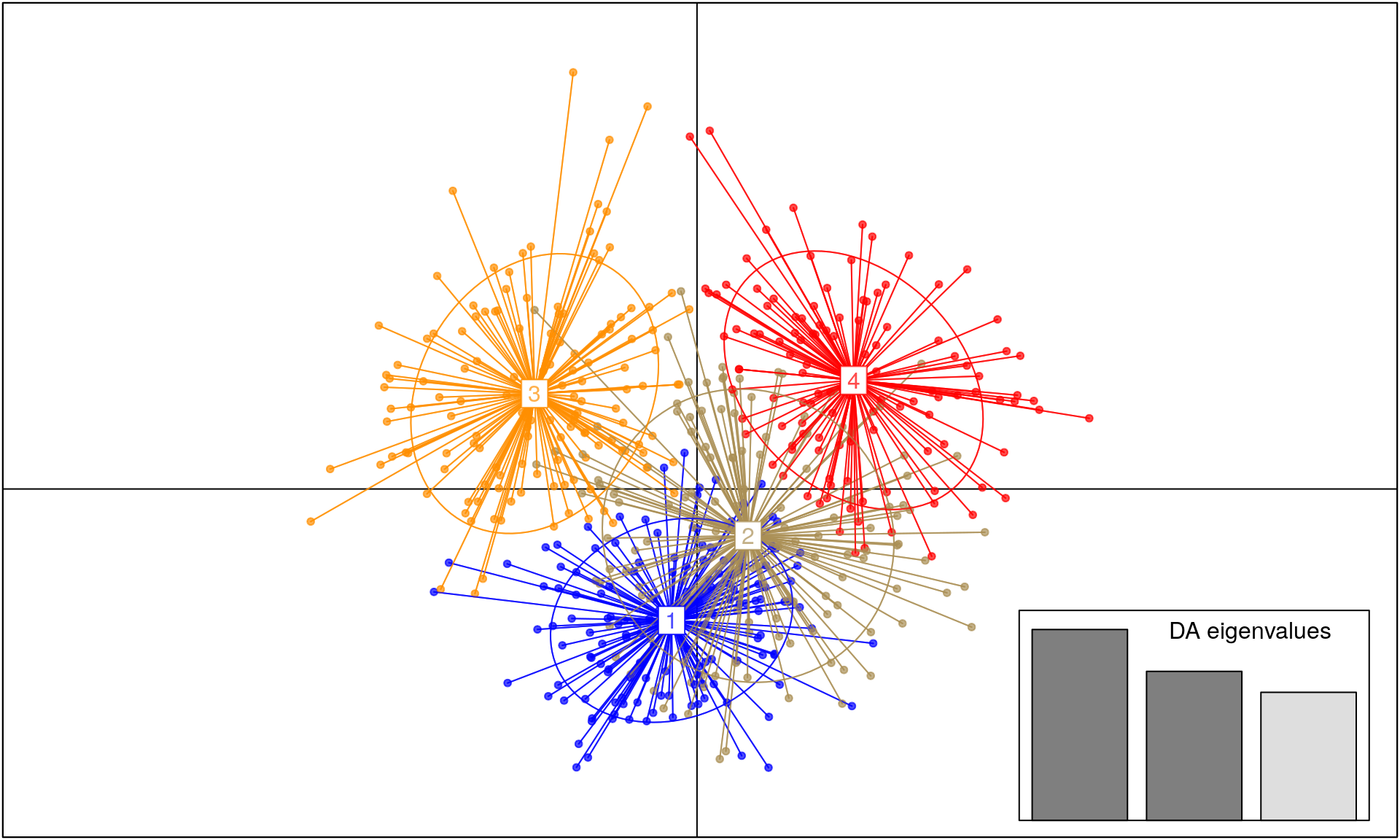
SambaR: An R package for fast, easy and reproducible population‐genetic analyses of biallelic SNP data sets - Jong - 2021 - Molecular Ecology Resources - Wiley Online Library

SambaR: An R package for fast, easy and reproducible population‐genetic analyses of biallelic SNP data sets - Jong - 2021 - Molecular Ecology Resources - Wiley Online Library
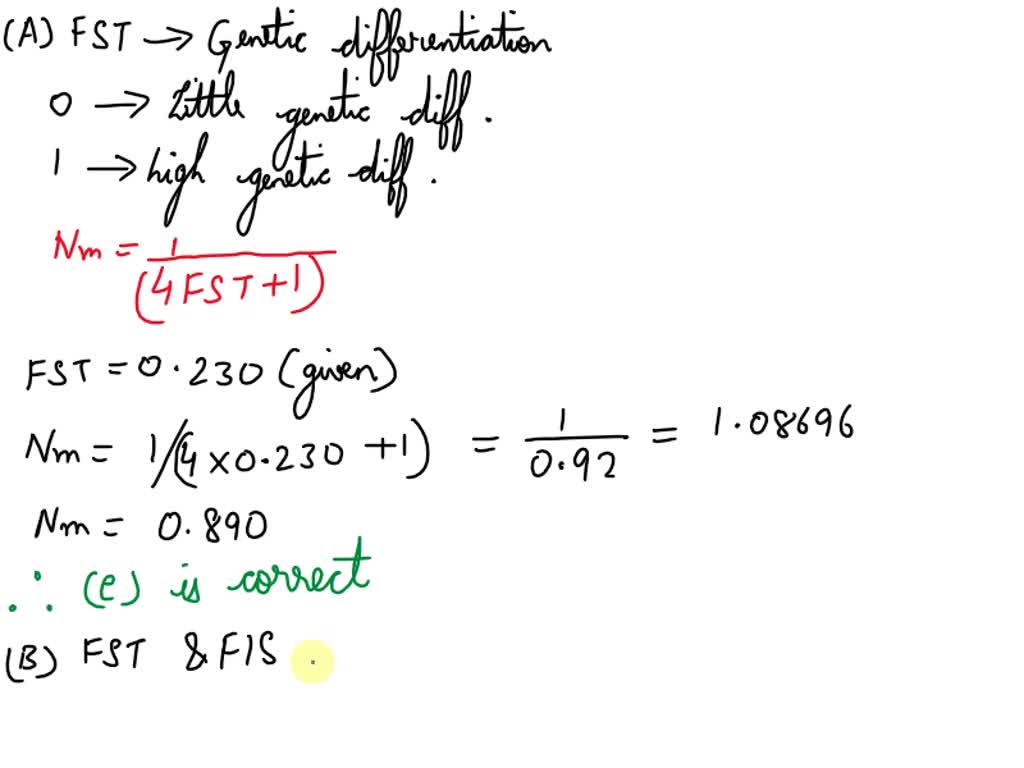
SOLVED: A) You calculate FST for two sub-populations of dung beetle, FST = 0.230. Based on this, how many effective migrants are there per generation? Select one: a. 4.35 b. none of
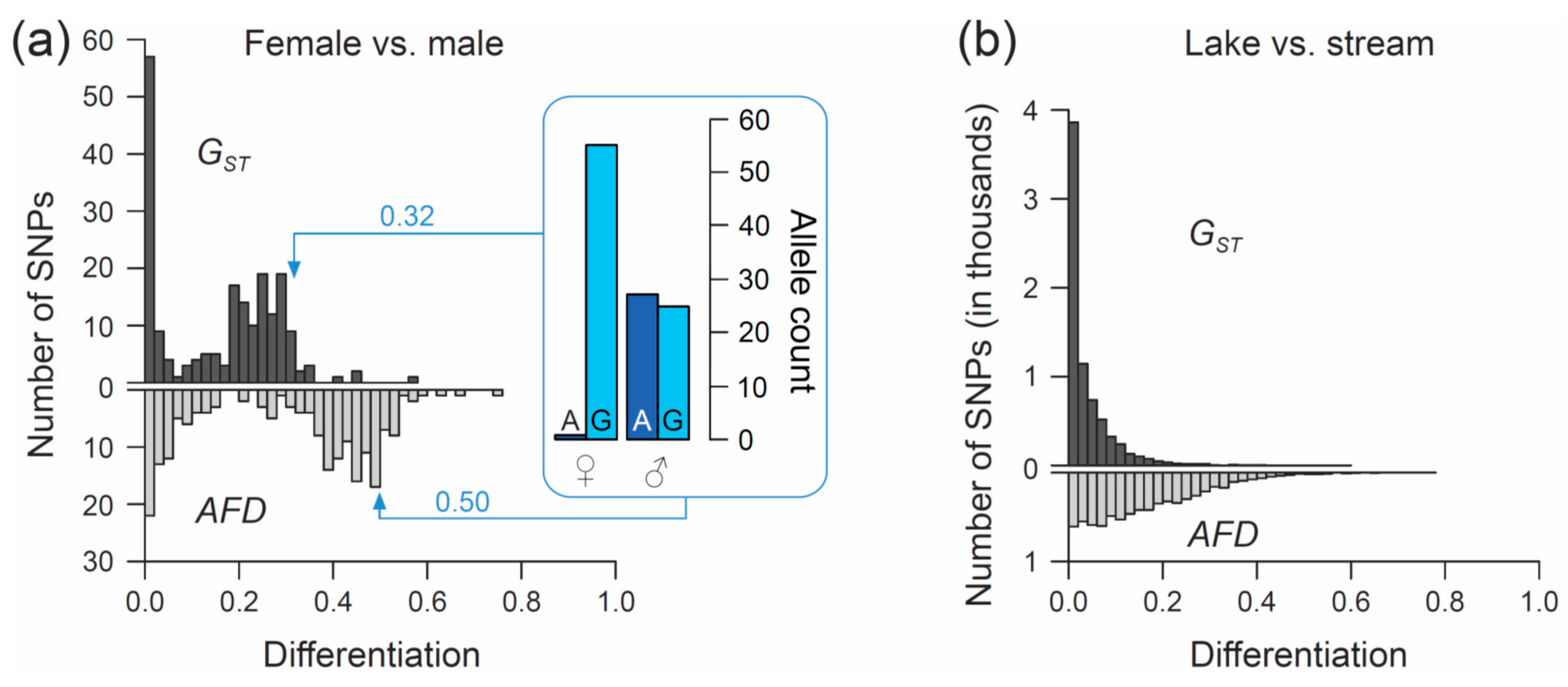
Genes | Free Full-Text | Allele Frequency Difference AFD–An Intuitive Alternative to FST for Quantifying Genetic Population Differentiation
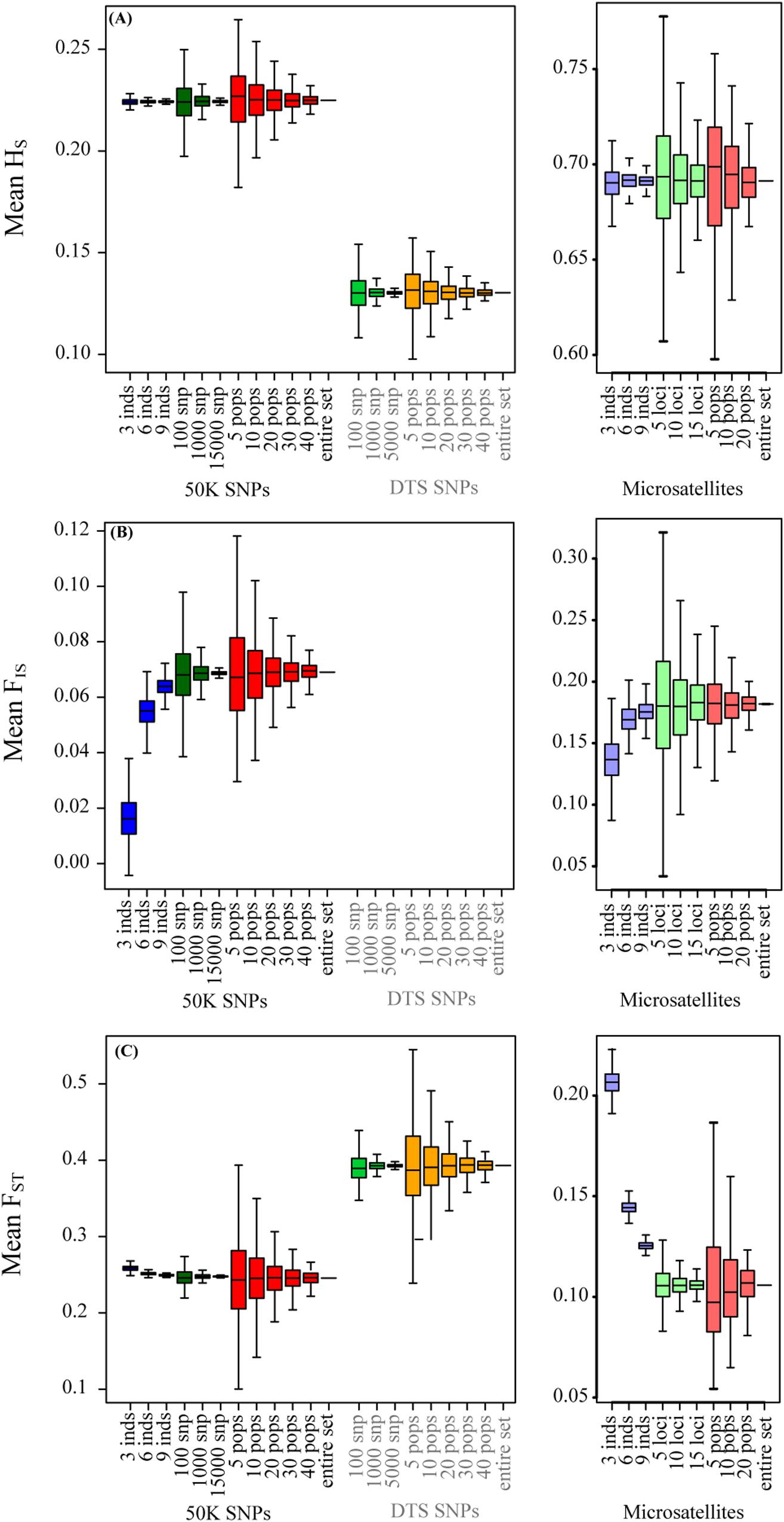
Frontiers | Evaluation of the Minimum Sampling Design for Population Genomic and Microsatellite Studies: An Analysis Based on Wild Maize
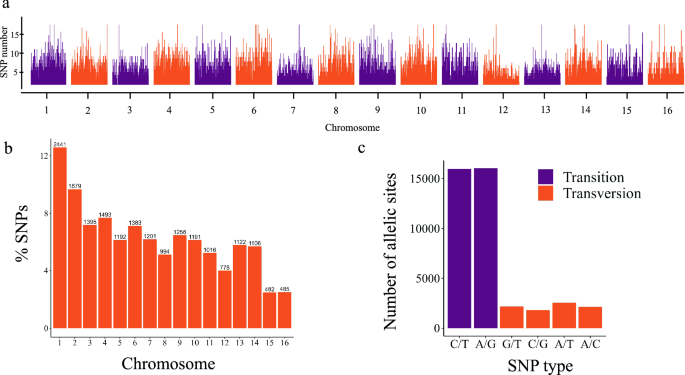
Genome-wide diversity analysis to infer population structure and linkage disequilibrium among Colombian coconut germplasm | Scientific Reports

FST values from locus-by-locus AMOVA. FST values were calculated at... | Download Scientific Diagram
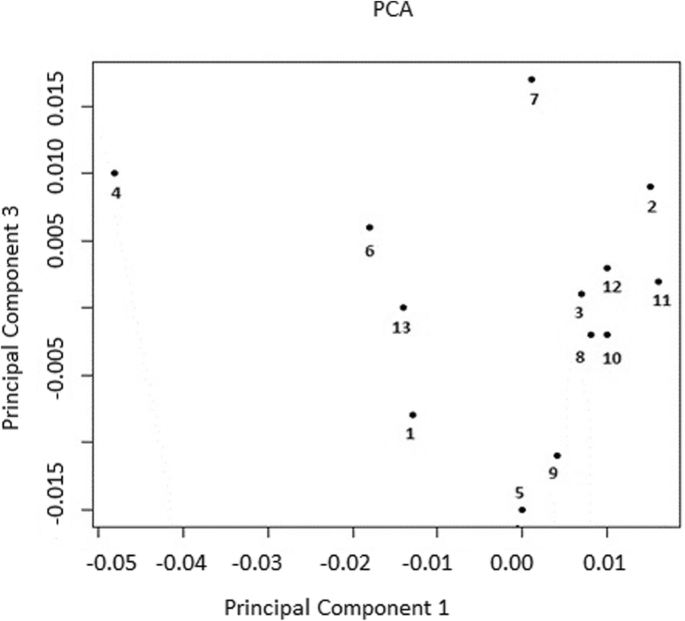
Assessing the power of principal components and wright's fixation index analyzes applied to reveal the genome-wide genetic differences between herds of Holstein cows | BMC Genomic Data | Full Text
Estimates of Genetic Differentiation Measured by FST Do Not Necessarily Require Large Sample Sizes When Using Many SNP Markers | PLOS ONE